Welcome to glmtools’s documentation!¶
import yaml
import numpy as np
import glmtools as glm
# Design config object contains methods for adding regressors, contrasts
# and ftests.
DC = glm.design.DesignConfig()
DC.add_regressor(name='Cond1',rtype='Categorical',codes=1)
DC.add_regressor(name='Cond2',rtype='Categorical',codes=2)
DC.add_regressor(name='Covariate', rtype='Parametric', datainfo='covariate', preproc='z')
DC.add_simple_contrasts()
for subj in range(1):
# Make a fake dataset
condition = np.tile( [1,2],4 )
session = np.repeat([1,2,3,4],2)
covariate = np.array([-3,-2,-1,0,0,1,2,3])
# Some random data
X = np.random.randn(8,)
# The data object must contain a 'data' kwarg pointing to the array to
# be analysed. Any other kwargs are considered to be meta-data and
# pushed into a dict in data.info
data = glm.data.TrialGLMData(data=X,
category_list=condition,
covariate=covariate,
session=session)
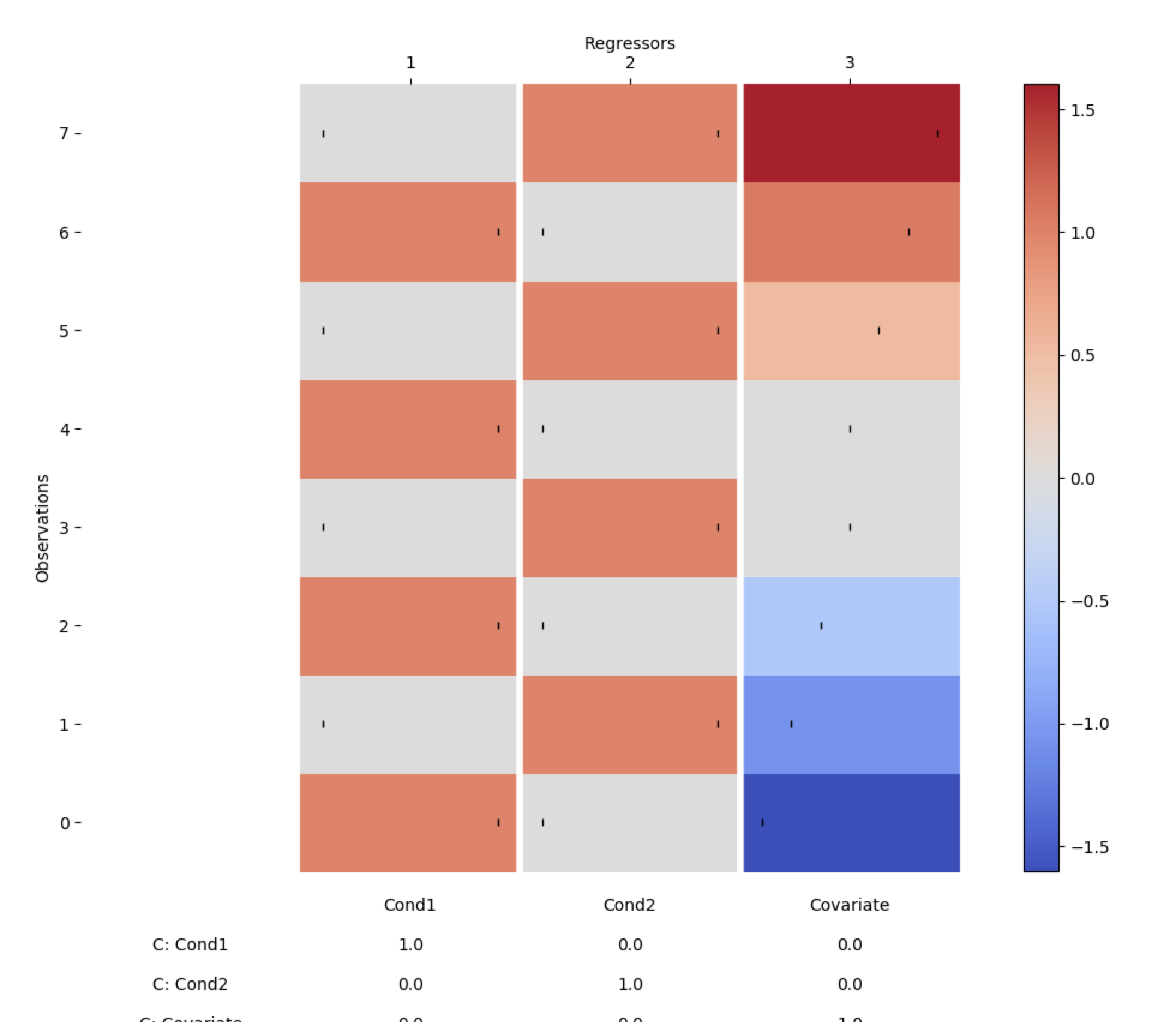
des = DC.design_from_datainfo(data.info)
des.plot_summary()
model = glm.fit.OLSModel(des,data)
Can also define DesignConfigs from yaml
yml = """
NewDesign:
regressors:
- {name: Cond1, rtype: Categorical, codes: 1}
- {name: Cond2, rtype: Categorical, codes: 2}
contrasts:
- {name: Cond1, values: 1 0}
- {name: Cond2, values: 0 1}
"""
DC = glm.design.DesignConfig(yaml_text=yml)
For contrasts in complicated designs we can specify regressors via a dict
y2 = """
NewDesign:
regressors:
- {name: Cond1, rtype: Categorical, codes: 1}
- {name: Cond2, rtype: Categorical, codes: 2}
- {name: Session, rtype: MeanEffects, datainfo: session}
contrasts:
- {name: Cond1, values: {Cond1: 1}}
- {name: Cond1, values: {Cond2: 1}}
- {name: CondDiff, values: {Cond1: 1, Cond2: -1}}
"""
DC = glm.design.DesignConfig(yaml_text=y2)
This can extend to some more complicated designs
Y = np.random.randn(40,8)
sessions = np.tile(np.arange(10),4)
subjs = np.repeat(np.arange(4),10)
covariate = np.random.randn(40,)
group_data = glm.data.TrialGLMData(data=Y,
sessions=sessions,
subjs=subjs,
covariate=covariate)
DC = glm.design.DesignConfig()
DC.add_regressor(name='Constant',rtype='Constant')
DC.add_regressor(name='Sessions',rtype='Parametric',datainfo='sessions',preproc='z')
DC.add_regressor(name='Covariate',rtype='Parametric',datainfo='covariate',preproc='z')
DC.add_regressor(name='Subj_{0}',rtype='MeanEffects',datainfo='subjs')
DC.add_contrast(name='GroupMean', values={'Constant':1})
DC.add_contrast(name='SessionEffect', values={'Sessions':1})
DC.add_contrast(name='CovariateEffect', values={'Covariate':1})
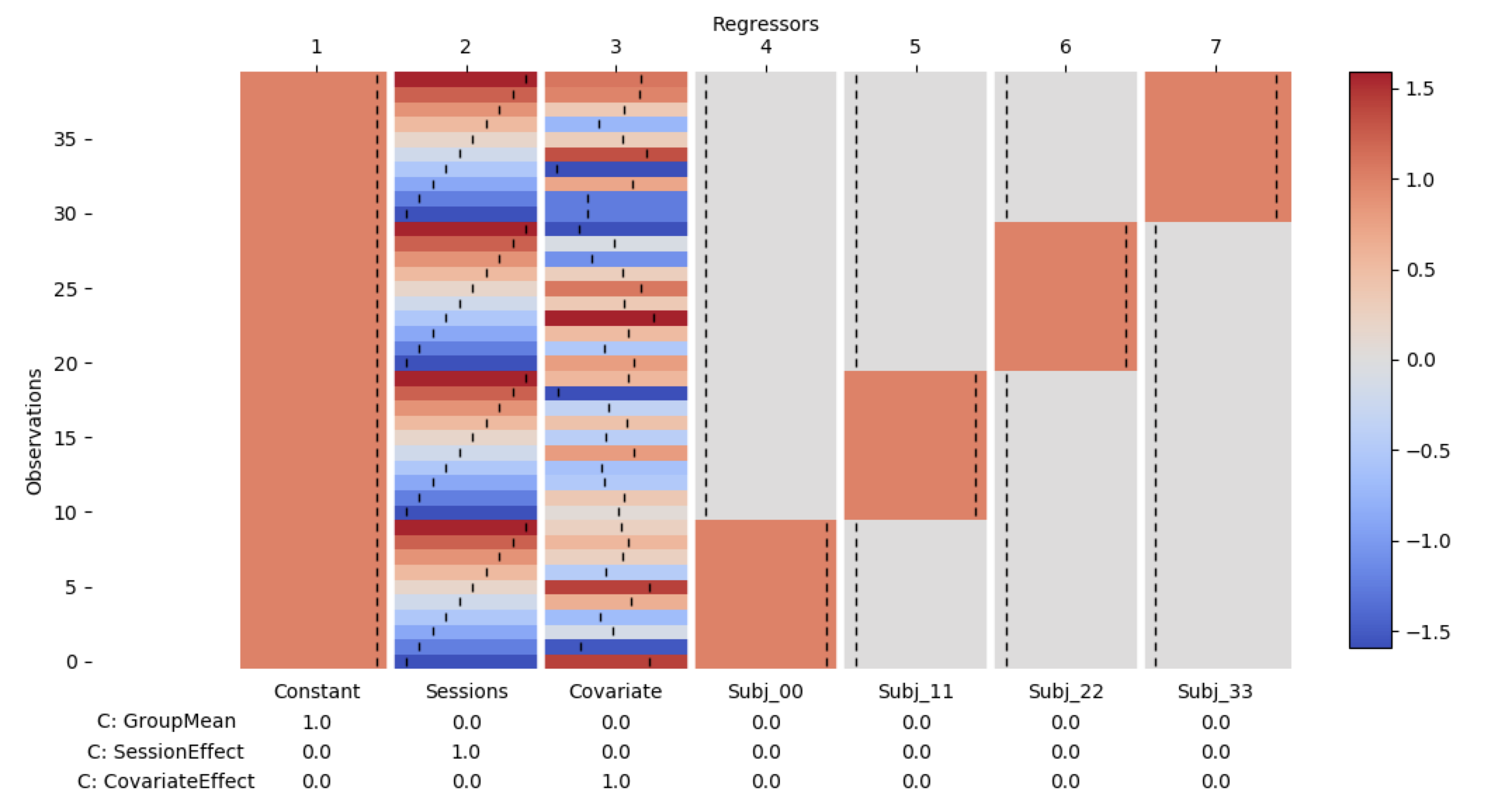
des = DC.design_from_datainfo(group_data.info)
des.plot_summary()
yml = """
NewDesign:
contrasts:
- name: GroupMean
values: {Constant: 1}
- name: SessionEffect
values: {Sessions: 1}
- name: CovariateEffect
values: {Covariate: 1}
regressors:
- {name: Constant, regressor: ConstantRegressor}
- {datainfo: sessions, name: Sessions, preproc: z, regressor: ParametricRegressor}
- {datainfo: covariate, name: Covariate, preproc: z, regressor: ParametricRegressor}
- {datainfo: subjs, name: 'Subj_{0}', regressor: MeanEffectsRegressor}
"""
DC = glm.design.DesignConfig(yaml_text=yml)
Some permutation features
# Permutations
P = glm.permutations.MaxStatPermutation(design, data, 1,
nperms,
metric='tstats',
perm_args=perm_args)
# 2d cluster permutations
perm_args = {'cluster_forming_threshold': 2.3, 'pooled_dims': (1, 2)}
CP = permutations.ClusterPermutation(design, data, cont,
500,
perm_args=perm_args,
metric='tstats')
cluster_masks, cluster_stats = CP.get_sig_clusters(data, 99)
Using sklearn model fits
# glmtools OLS model
model = glm.fit.OLSModel(design,data)
# sklearn OLS model
skmodel1 = glm.fit.SKLModel(design,data)
# sklearn robust regression
skmodel2 = glm.fit.SKLModel(design,data,fit_args={'lm':'RANSACRegressor'})